Single Molecule Detection and Analysis Methodology
In single molecule optical tracking experiments, photophysical and kinetic differences between different analytical targets are often small. This obscures species identification and prohibits analyses in complex media, such as lipids and cell membranes. We are exploring the use of pattern recognition tools to classify trajectories of single molecules diffusing in 2D. Here, Principal Component Analysis is coupled with SIMCA to successfully identify two species with overlapping emission spectra, but different translational and photophysical kinetics. One species (pink) arises from fluorescently labeled lipids diffusing in the membrane. A second species (green) arises from fluorescent contaminants that adhere to the substrate from solution.
Confocal fluorescence microscopy, traditionally used for 3D imaging purposes, can be adapted for single-molecule detection capability. In this arrangement ultrasensitive measurements of molecular motion can be acquired as molecules diffuse in and out of a tightly focused confocal laser beam. Frequently, the raw intensity vs. time data record is processed via fluorescence correlation spectroscopy. Our group is actively involved in broadening this techniques for a variety of chemical and biological applications, including analyses inside cells, cell membranes, and also in conjunction with biological nanopores.
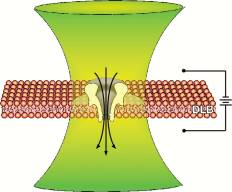 Combining the instrumentation for single-channel electrical recording with single-molecule fluorescence microscopy provides a means for simultaneously interrogating channels with light and electricity in real time. The combined approach creates an information-rich technique for the basic study of channel dynamics, molecular transport, and intercellular communication.
Combining the instrumentation for single-channel electrical recording with single-molecule fluorescence microscopy provides a means for simultaneously interrogating channels with light and electricity in real time. The combined approach creates an information-rich technique for the basic study of channel dynamics, molecular transport, and intercellular communication.

